How To Collect NMR Spectra for Optimal Performace with Bayesil
In order for Bayesil to perform optimally, it is best if the sample preparation and the NMR data acquisition are performed in a manner similar to that used to prepare and collect the reference NMR spectra used in Bayesil's spectral library. Using different spectral preparation and collection conditions will compromise the performance, particularly the quantification accuracy. Here we describe the recommended methods that Bayesil users should follow when preparing biological samples and collecting NMR spectra of those samples.
Sample Preparation
All samples should have been either stored frozen or stored at 4 °C prior to use. Cerebrospinal fluid should be spun down to remove particulate matter and its pH checked (or adjusted) to ensure that it is between pH 6.8-7.4. More attention must be paid for the sample preparation of serum or plasma because of the high concentration of proteins in these samples. Proteins in the biofluid sample of interest must be removed by passing it through a pre-rinsed (washed 7X) Amicon Ultra-0.5 3000 MWCO filter at 4 °C using a centrifuge (12,000 g force). Unwanted protein signals are particularly problematic with serum and plasma and ultrafiltration is currently the best method of removing these signals without altering the chemical composition of the biofluid. After the protein filtration step has been completed, the sample may be placed into a NMR tube. If Shegemi tubes are used, 285 µL of sample will be needed. For regular thin walled 5 mm NMR tubes, 570 µL of sample will be needed. With respect to pre-rinsing the Amicon filters, sterile double-distilled water or its equivalent should be used. Rinsing should be done at least seven times to ensure any residual glycerol is removed. Otherwise glycerol concentrations will be erroneously elevated in the biofluid sample.
It is recommended that users check the pH of each sample prior to spectrum collection to ensure the pH is near 7.0. Otherwise the global fitting routine will produce larger errors than reported. The physiologic pH of blood and CSF is between 7.3 and 7.4. Plasma and serum generally have large buffering capacities, thus pH adjustment is rarely problematic. Bayesil is designed to work well with samples having a pH range between 6.8 and 7.5. However, the pH may vary due to sample handling or disease state and adequate optimization of the pH prior to data collection cannot be stressed enough. Larger concentrations of sodium phosphate than 50 mM may be necessary if your sample's pH is not adequately controlled. Thus, checking your sample's pH beforehand can alleviate many potential problems.
The necessary volumes and buffer recipes required for sample preparation are shown below in Table 1 and Table 2 respectively.
Table 1: Sample components.
| Sample Size | Biofluid Volume | D2O | Buffer | Final Volume | NMR tube |
|---|---|---|---|---|---|
| 350 µl | 285 µl | 35 µl | 30 µl | 350 µl | Shegemi |
| 700 µl | 570 µl | 70 µl | 60 µl | 700 µl | Regular 5 mm |
Table 2: 11.7x Buffer Recipe.
| Ingredients | 11.7x Stock (100 ml) | Concentration | Final Concentration |
|---|---|---|---|
| Na2HPO41 (anhydrous) | 7.850 g | 583 mM | 50 mM |
| DSS2 | 0.230 g | 11.7 mM | 1.0 mM |
Table 3: Alternate 11.7x Buffer Recipe.
| Ingredients | 11.7x Stock (100 ml) | Concentration | Final Concentration |
|---|---|---|---|
| K2HPO41 (anhydrous) | 30.450 g | 1.75 M | 150 mM |
Sodium Formate2 or 2-chloro pyrimidine-5-carboxylic acid2 | 0.0079 g or 0.0926 g | 1.167 mM or 5.84 mM | 100 uM or 500 uM |
| DSS-d63 | 0.115 g | 5.833 mM | 0.5 mM |
NMR Spectral Acquisition Parameters
All one-dimensional 1H-NMR spectra should be collected using the first increment of a standard two-dimensional presaturation NOESY experiment (see pulse sequence below).
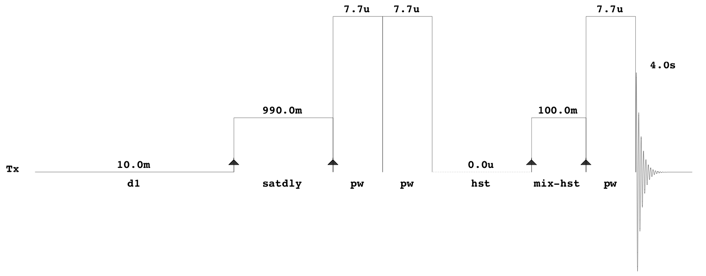
With Agilent/Varian instruments, this experiment can be found in the BioPack experimental library and has the pulse sequence label “tnnoesy”. The comparable Bruker experiment is CPPWaterSupp (pulse program: pusenoesypr1d). All NMR spectra should be collected at 25 °C. A 12 ppm sweep width, a 4 s acquisition time, a 100 ms mixing time, a 10 ms recycle delay and a 990 ms saturation delay should be used. Typically, 128 transients should be acquired for samples collected at 500 MHz or 600 MHz, although this may vary depending on the quality of the probe (cold/cyroprobe versus room temperature probe). Eight steady-state scans should collected prior to data acquisition and the pre-saturation pulse power should be calibrated to provide a field strength no greater than 60 to 80 Hz. This minimizes the saturation of signals near the water resonance (4.78 ppm). In addition to the presaturation pulse calibration, both the transmitter offset and the saturation pulse should be positioned on the water resonance. No other solvent suppression pulses, including gradients, should be used (i.e. all gradient pulse widths and powers should be set to zero). Standard phase cycling should be used to suppress the water resonance and other spectral artifacts. Thus, it is imperative that the pulse sequence be set up such that the number of transients is a multiple of the number of pulses in each cycle. For the Varian/Agilent sequence, it is 16. All excitation pulses should be calibrated to correspond to a 90° flip angle (7.7 µs for the probe used in the above illustrated pulse sequence).