Welcome to Bayesil
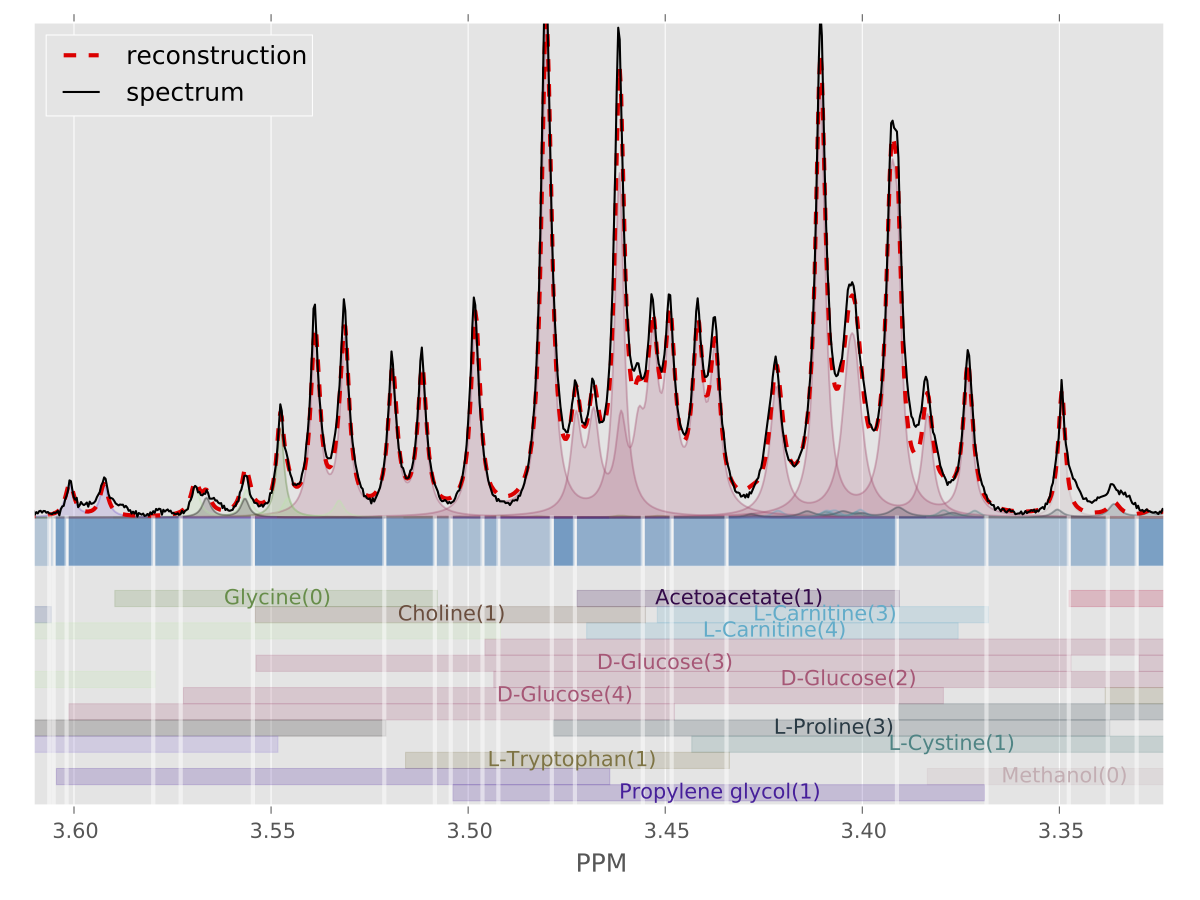
Bayesil is a web system that automatically identifies and quantifies metabolites using 1D 1H NMR spectra of ultra-filtered plasma, serum or cerebrospinal fluid. The NMR spectra must be collected in a standardized fashion (see How To Collect NMR Spectra for Bayesil) for Bayesil to perform optimally. Bayesil first performs all spectral processing steps, including Fourier transformation, phasing, solvent filtering, chemical shift referencing, baseline correction and reference line shape convolution automatically. It then deconvolutes the resulting NMR spectrum using a reference spectral library, which here contains the signatures of more than 60 metabolites (see here for a list). This deconvolution process determines both the identity and quantity of the compounds in the biofluid mixture. Extensive testing shows that Bayesil meets or exceeds the performance of highly trained human experts.
Citing Bayesil:

NMR Metabolomics Analysis Kit
Serum and CSF
• For preparation of 50 serum or CSF samples
• Processing time: 2 min/sample
• Access to Bayesil batch processing feature
• Process and analyse samples for as low as $10/sample
More info hereClick to order